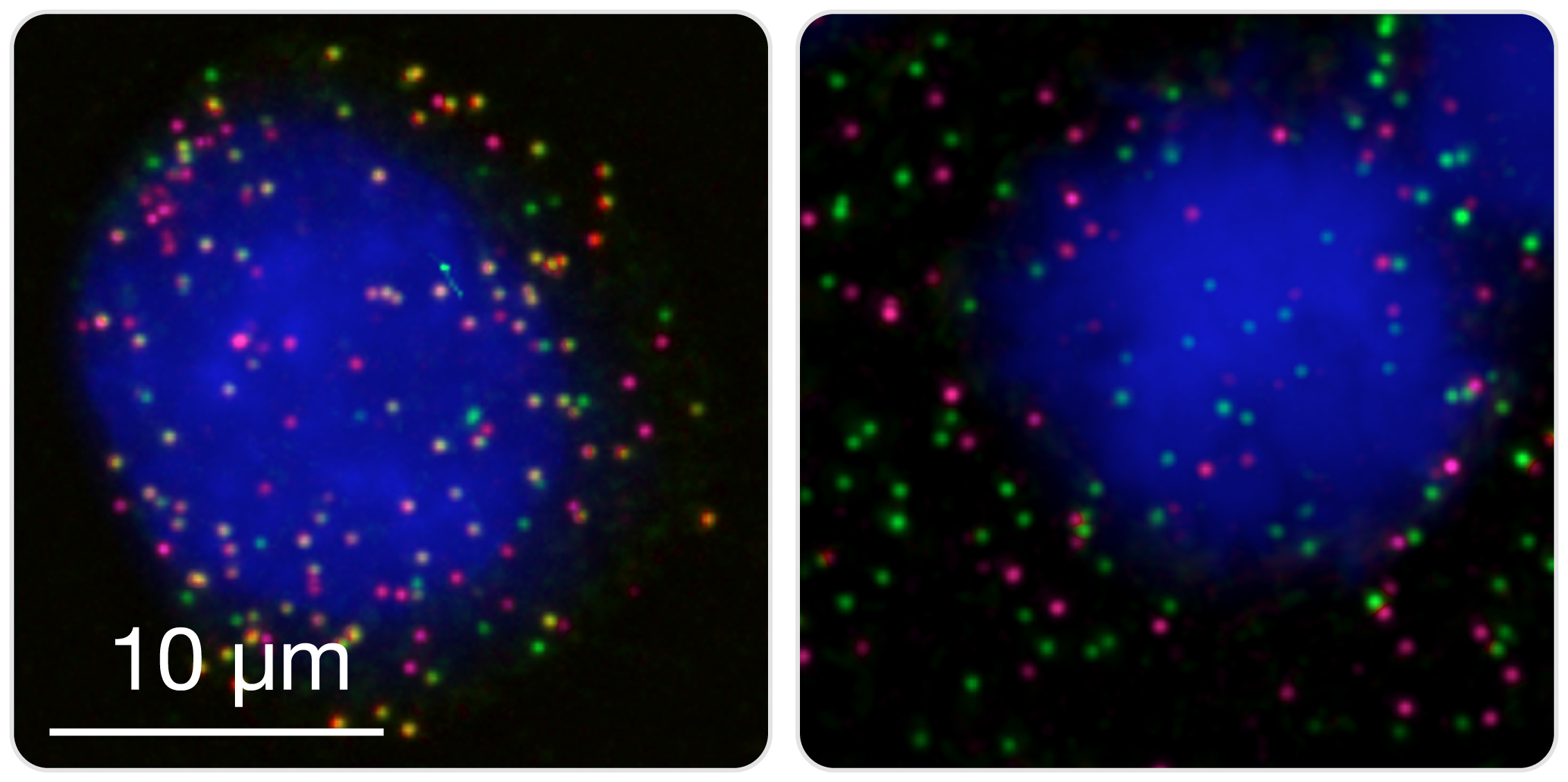
Detect and count fusions in cultured cells or cytologic specimens. Cultured cell lines or primary cells, as well as cells directly extracted from patients (for example, white blood cells from peripheral blood or bone marrow) are fixed in a methanol-acetic acid solution mix and spotted on microscopy slides (manually or by cytospin). Fused partners are then hybridized with two differently labeled smFISH probe sets, and appear as colocalized diffraction-limited spots in the nucleus and cytoplasm. Left image: K562 cell expressing BCR (green) and ABL1 (magenta) transcripts. Multiple spatially colocalized spots (yellow) corresponding to BCR-ABL1 transcripts can be seen. Right image: TS cell expressing BCR (green) and ABL1 (magenta) transcripts, but not BCR-ABL1.
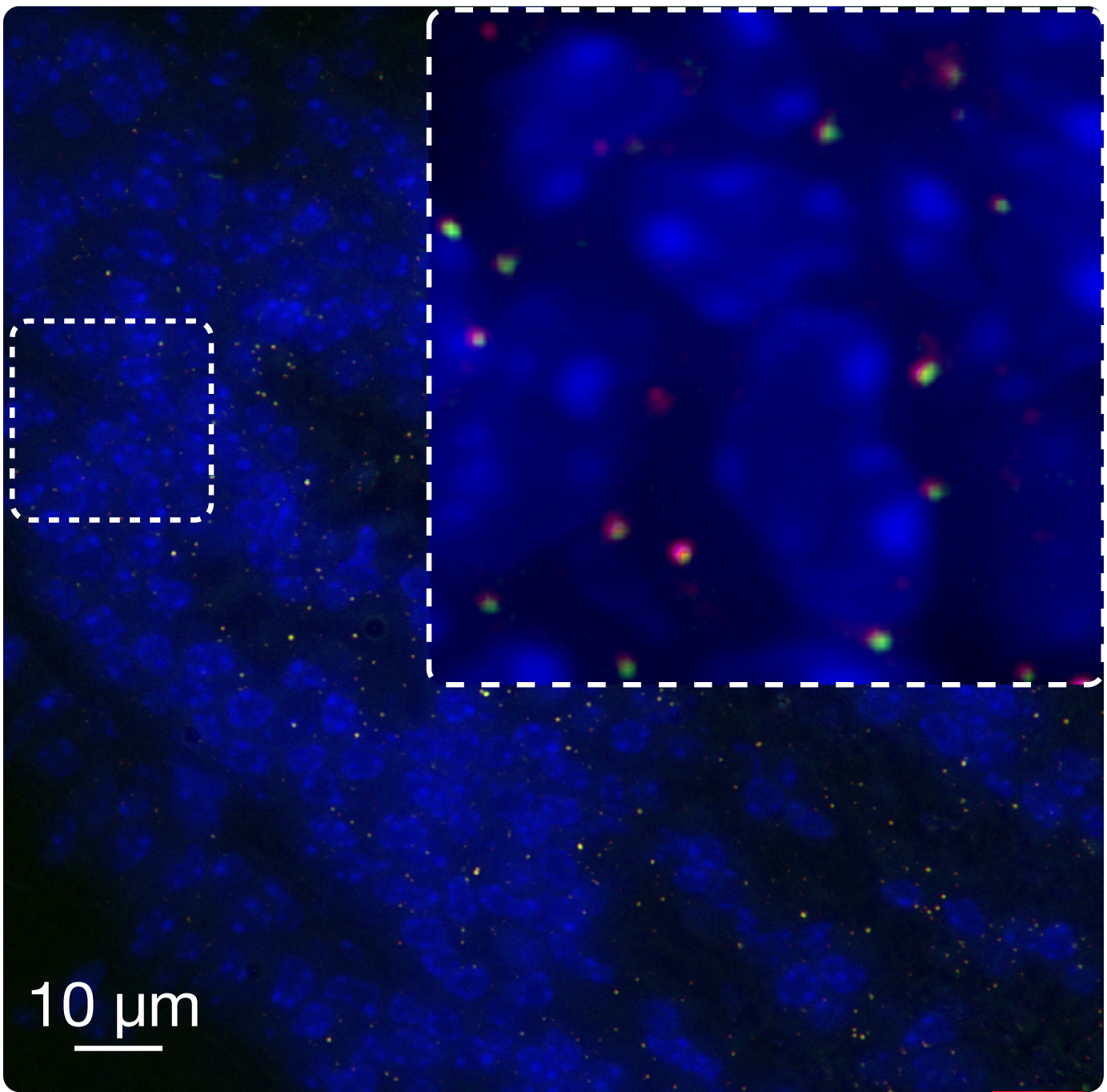
Diagnose fused transcripts in tissue sections. Both frozen and formalin-fixed, paraffin-embedded (FFPE) tissue sections can be used to detect fused transcripts using FuseFISH. FFPE sections are first deparaffinized, and RNA retrieval and autofluorescence quenching are performed (see Downloads, Protocols). Fused partners are then hybridized with two differently labeled smFISH probe sets, and appear as colocalized diffraction-limited spots. In the image, EML4-ALK fused transcripts (yellow spots) are detected in an FFPE section of a mouse xenografts of human Non Small Cell Lung Cancer.
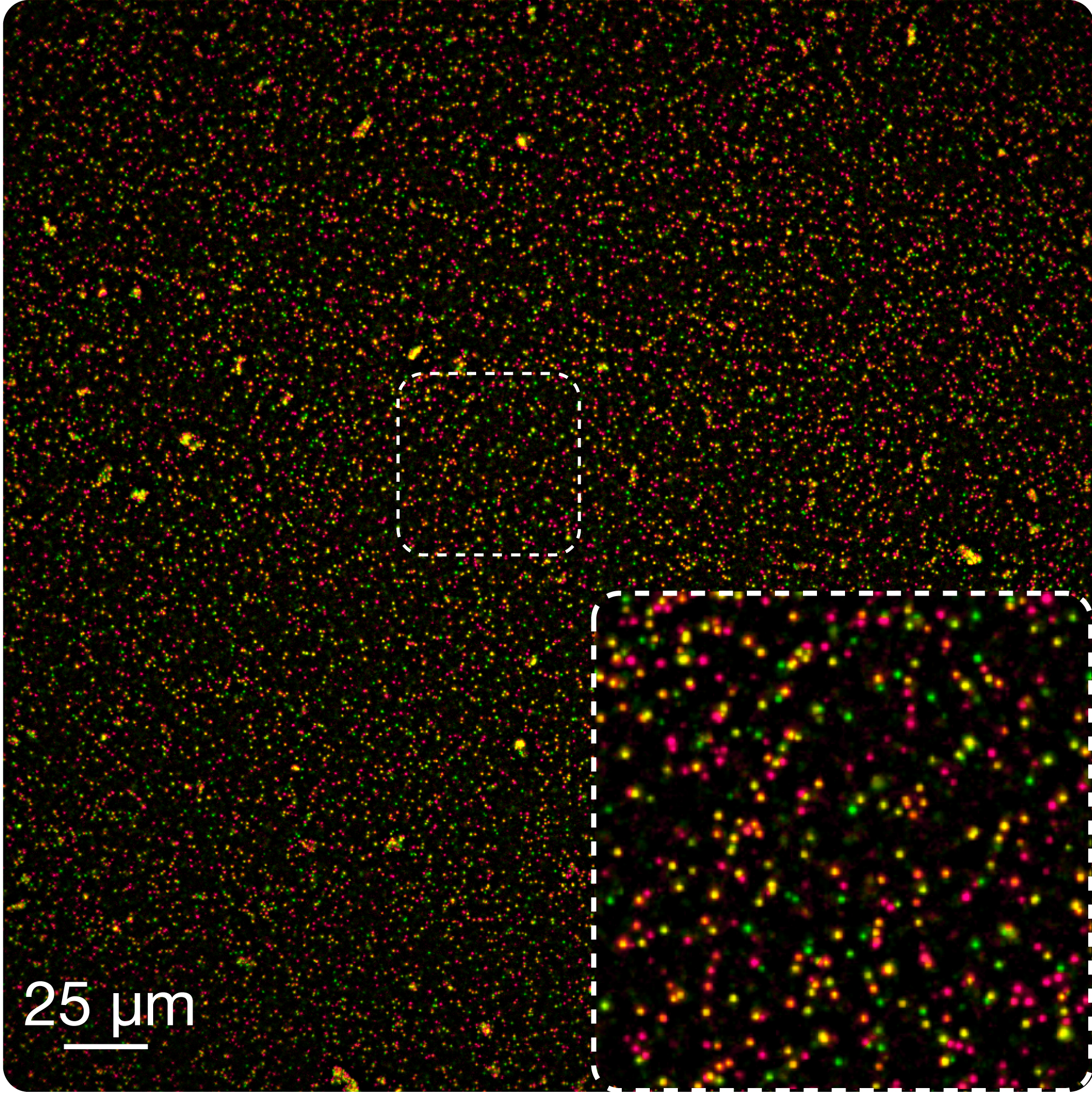
Find transcript fusions in purified RNA. A mixture of proteinase K and purified RNA is dried onto a glass support (microscope slide or coverglass) and fixed with a 3:1 vol./vol. mixture of methanol and acetic acid. A stable RNA-protein matrix is formed, which can be then rapidly hybridized with smFISH probes. Fused partners are visualized using two differently labeled smFISH probe sets, and appear as colocalized diffraction-limited spots. This application is particularly suitable for rapid validation of fusions newly discovered by next-generation sequencing using leftover RNA. Combination of multiple hybridization-imaging-probe stripping cycles on a high-throughput imaging platform allows, in principle, multiple fusions to be detected in the same experiment run (multiplexing) in multiple samples (high-throughput).